Preparing external biomet files
These instructions describe how to prepare biomet files that were collected on a separate datalogger or saved in a separate data file from your fluxes (not in LI-COR .ghg file format). There are two options to consider.
The first option is to use EddyPro desktop software to process or reprocess your raw data. If your raw flux data was collected or stored on a LI-COR system but your slow biomet data was collected on a separate datalogger, you should still have .ghg files with two raw CSV files. See (1) and (2) in Figure 2‑1. Now you can use your separately collected (external) biomet files during flux computations (3), as shown in Figure 2‑1.
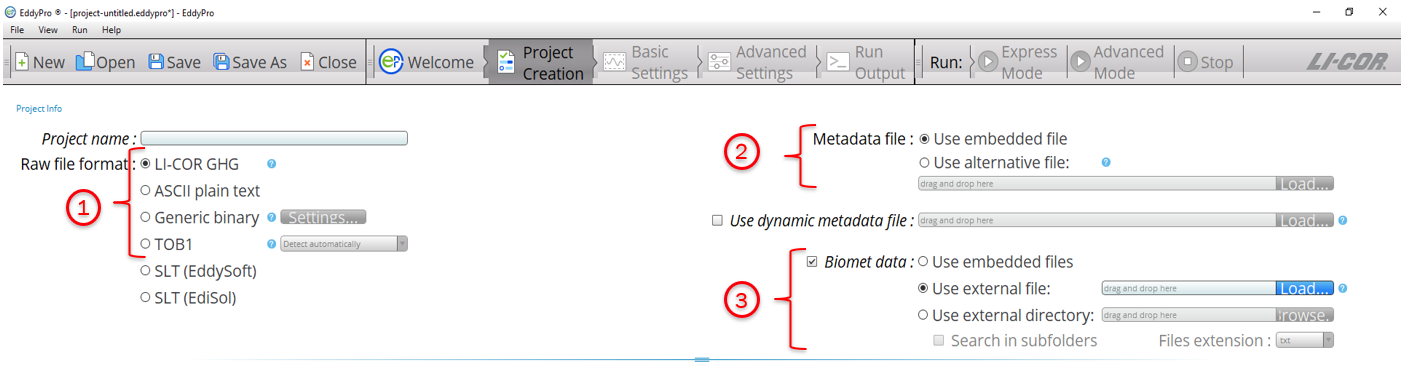
Note: If your raw flux data was collected or stored on a datalogger, then you will need to choose either “TOB1” or “ASCII” in step 1 and go through the ‘Metadata File Editor’ section in EddyPro to describe your data.
Before loading the external Biomet file, be sure it is in the correct format for EddyPro to read (variable names, units, etc.). Instructions for preparing the external biomet file are in EddyPro help at Supported external file formats.
Run EddyPro. In the output folder, you will now have formatted biomet files that you can directly import and use in Tovi.
Benefits to this first option are that your flux computations may improve as the appropriate biomet variables are used in some of the algorithms and corrections. You will also have both the resulting ‘full-output’ and ‘biomet’ files to import directly into Tovi in the correct formats. The drawback is the time it may take to reprocess your data (based on number of years, for example, this may take overnight) and if your results are different from the first time you processed, based on either settings used or the inclusion of the biomet files.
The second option is to simply restructure your existing ‘Biomet output file’ similar to what EddyPro would produce for a biomet output file. It involves a few key steps.
- The file needs to be formatted similar to an EddyPro biomet output file with one line/row for variable labels followed by another line/row with units.
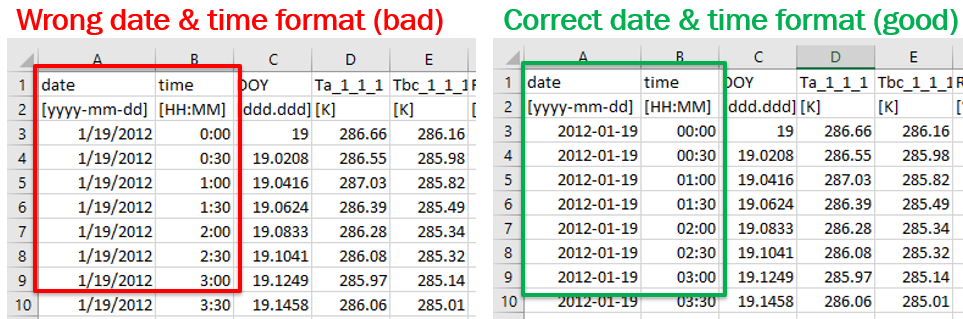
- The first three columns of the biomet file need to be ‘date’, ‘time’ and ‘DOY’ and it is important that these each need to be in the correct format. See the image below.
- The timestep should be 30 minutes.
- The starting and ending dates and times in your biomet data file must match those in the EddyPro flux ‘full-output’ file to appropriately integrate with the simultaneous flux records. Check the start and end times/dates of both files. If one of these files is longer than the other, you may want to trim down the start/end date/time so they are the same.
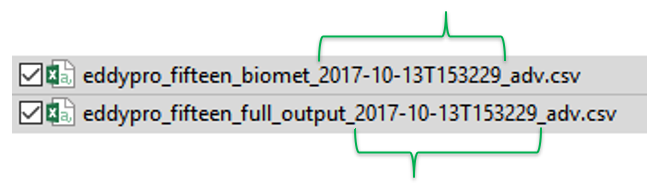
- Similarly, during importing files, Tovi groups and synchronizes the EddyPro output files and the biomet output files based on their file name. So the biomet file name structure must be similar to the full output file (see the image below). Notice how the date and time in the file name match between the two files.
- The variables and units need to match what EddyPro would produce in a biomet output file. Again, reference the instructions in EddyPro help at Supported external file formats.
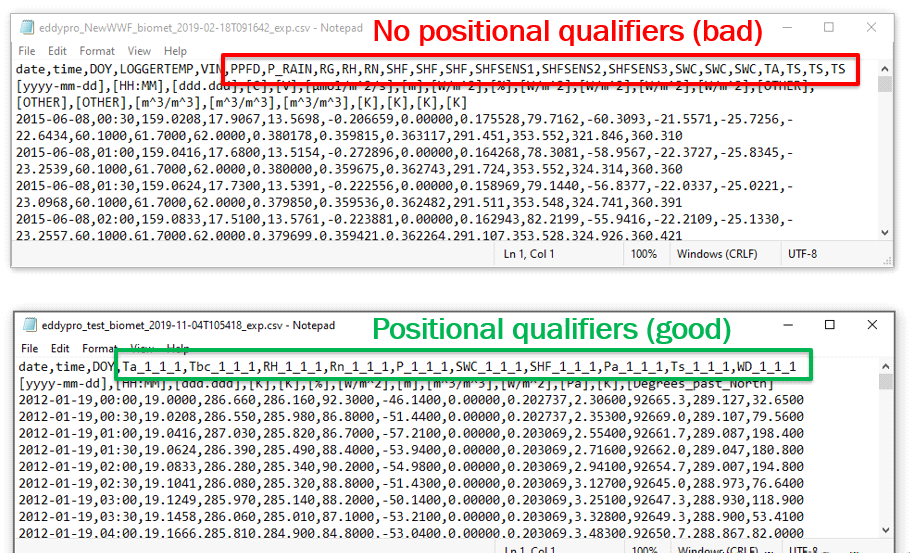
- Biomet variables need the ‘positional qualifiers’ included in the variable names. This is the “_H_V_R” as described at the two links below:
- http://www.europe-fluxdata.eu/home/guidelines/how-to-submit-data/variables-codes
- https://ameriflux.lbl.gov/data/aboutdata/data-variables/#base (scroll halfway down the page) with some example shown here: https://ameriflux.lbl.gov/data/data-variable-qualifier-examples/ .
- This follows the new FLUXNET (ICOS/AmeriFlux) standards for variable naming for submission to their networks. EddyPro automatically appends them on the biomet outputs if they are not originally included. So, for your independent biomet variables, you will want to have the variable labeled, for example, as: TA_1_1_1, TA_2_1_1, NETRAD_2_1_1, etc. (Without the positional qualifiers, Tovi cannot differentiate TA from the Analyzer with TA from the biomet system.) You can make these edits in a program like Notepad (see below).
- It is not necessary to do this for every biomet variable that you have. Below are the biomet variables that can or will be used inside Tovi (other variables will come from the EddyPro ‘full-output’ file). If you have any of the variables in the list below, you can make sure they are named properly, have the correct units, and apply the positional qualifiers.
- Meteorology variables: TA, PA, P_RAIN, RH, WS, WD, VPD, CO2, CH4
- Radiation variables: SW_IN, SW_DIR, SW_DIF, SW_OUT, LW_IN, LW_OUT, NETRAD, PPFD_IN, PPFD_DIR, PPFD_DIF, PPFD_OUT
- Soil variables: SHF, TS, SWC, WATER_TABLE_DEPTH
- Turbulence variables: MWS, WS
- Water variables: H2O, RH, P, P_RAIN, P_SNOW, SWC, WATER_TABLE_DEPTH
- If you have biomet variables besides those listed above, you can reformat them as you wish or by what is required for network submission purposes.
Important note if you pursue this second option:
Often when editing files in Excel®, the date format gets modified automatically inside the spreadsheet software to a format that Tovi will not recognize when importing the file (for example, 2010/02/11). Similarly, Excel may convert your time to three digits instead of four (for example, 1:30), which Tovi will not recognize. You will want to make sure the files are saved with date in the format of ‘yyyy-mm-dd’ and time in the format of ‘hh:mm’ as shown in step #2 above).