Gene Knockdown and Knockout Confirmation
Knockdown and knockout are two methods used to silence a target gene of interest. The partial (knockdown) or complete (knockout) silencing of a target gene enables researchers to identify and validate biological targets of human health significance. These techniques also enable researchers to better understand disease processes, create negative controls, and advance therapeutic discovery.
Quantitative Western blotting and the In-Cell Western™ Assay can be used to confirm the effects of gene knockdown/knockout. Both assays can be performed with exceptional quality and consistency using Odyssey® Imagers, such as the Odyssey M.
Confirming Target Knockdown with RNAi Analysis
RNA interference (RNAi) is a naturally occurring process to inhibit, or knockdown, the normal expression of a gene by deactivating or suppressing mRNA. RNAi can be artificially replicated in the laboratory using an RNAi vehicle, such as siRNA, shRNA, or other RNA-producing vector constructs. This allows for experimentation with a gene of interest. Knockdown of a specific gene allows researchers to analyze genetic function or to identify targets with potential therapeutic applications.
RNAi Analysis Using Quantitative Western Blotting
Quantitative Western blots are an essential tool for RNAi analysis. With quantitative Western blots, you can:
- Compare data from different vectors and constructs, such as siRNA, shRNA, or other RNA-producing vector construct
- Confirm the impact of your RNAi strategy on protein levels
- Analyze proteins in different signaling pathways to determine the impact silencing has on a phenotype, such as cell proliferation or migration


RNAi Analysis Using In-Cell Western Assay
In-Cell Western Assays can be used in a functional siRNA screen to measure knockdown effects in cultured cells. In-Cell Western Assays offer higher throughput for complex studies, as well as exceptionally consistent data (Z' factor).1


Hoffmann, et al. have demonstrated the In-Cell Western Assay can be used as a powerful cellular assay for genome-wide RNAi screens.2 In-Cell Western screening was used to assess the effects of knockdowns on mTORC1-dependent phosphorylation of ribosomal protein S6 (rpS6).2 Their research showed In-Cell Western RNAi screening to be faster and less expensive than high-content immunofluorescent microscopy while providing similar or better statistical replicability.




Confirming Target Knockout with CRISPR Analysis
CRISPR editing can be used to knockout a gene to inhibit the expression of certain proteins. Unlike knockdown, which inhibits a gene, knockout inactivates or removes a target gene completely. Like knockdown, knockout can be used to study the behavior and effects of a gene of interest to assess its function or its potential to affect human health.
CRISPR Analysis Using Quantitative Western Blotting
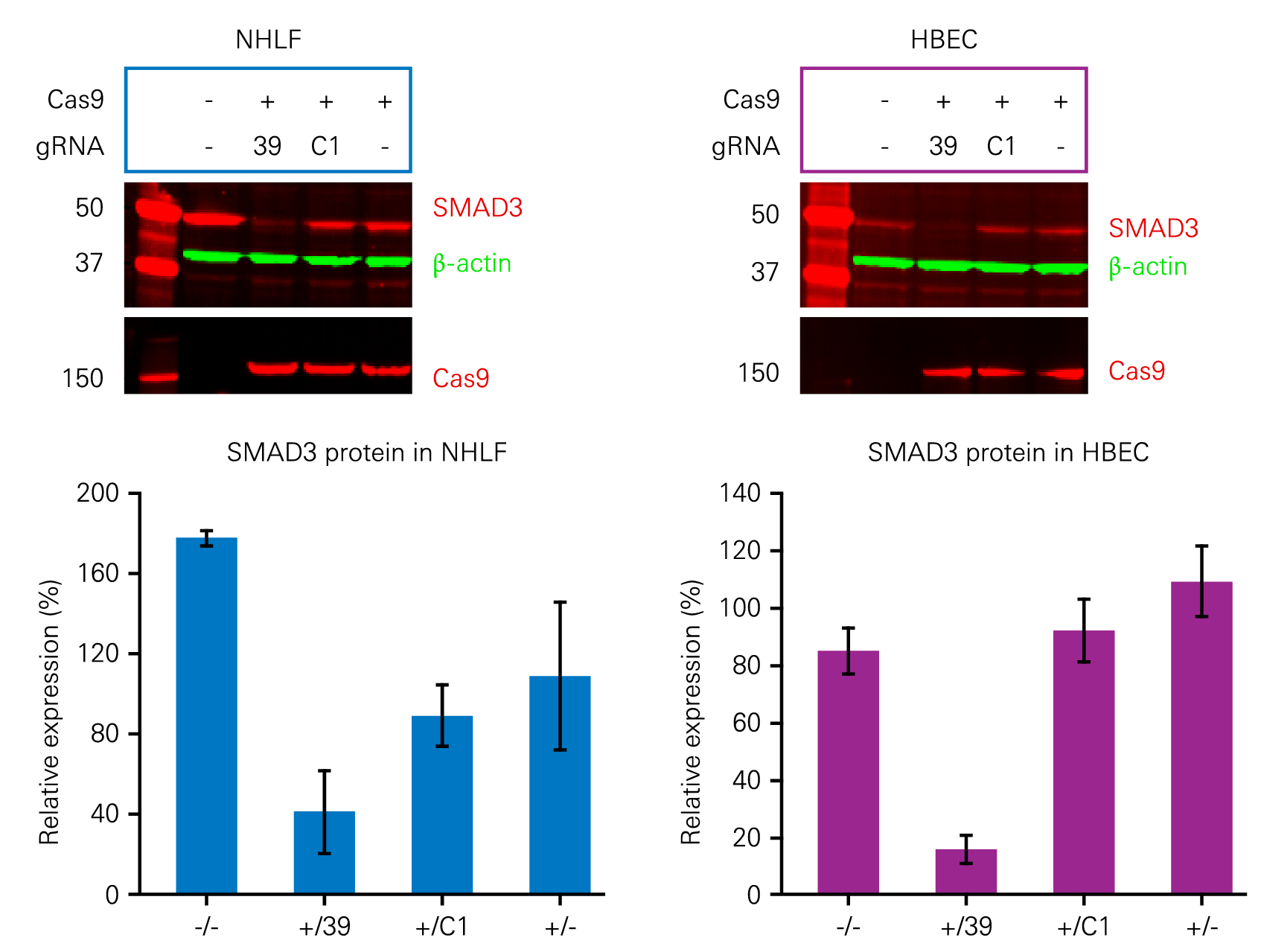
The effects of gene knockout via CRISPR can be confirmed using quantitative Western blotting. Figure 5 demonstrates the use of Adenoviral (AdV) CRISPR/Cas9 to inhibit the expression of SMAD3, a protein involved in triggering pulmonary fibrosis.
CRISPR Analysis Using In-Cell Western Assay
The In-Cell Western Assay can be used to measure the effect of knockout in cultured cells. Figure 6 shows the efficiency of ERK1 knockout when compared to wild-type HeLa cells.


What steps can you take today toward improved knockdown/knockout confirmation?
LI-COR provides products, protocols, and support for Western blotting, In-Cell Western Assays, and many other assays that help reduce variability and increase confidence in your results. Want to learn more? Contact LI-COR today and let us help you improve your knockdown/knockout confirmation.
Resources
- Revert™ Total Protein Stain Normalization Protocol
- Housekeeping Protein Validation Protocol
- Determining the Linear Range for Quantitative Western Blot Detection Protocol
- Housekeeping Protein Normalization Protocol
- Pan/Phospho Analysis for Western Blot Normalization Protocol
- Quantitative Western Blot Analysis with Replicate Samples Protocol
- Western Blot Normalization Handbook
- The Tactical Guide on Normalization eBook
- The Essential Guide to Successful Western Blots eBook
Protocols
Handbooks
eBooks
References
- Boveia, V., Ambroz, K.L.H., and Olive, D.M. (2009). Using the Z’-Factor Coefficient to Monitor Quality of Near-Infrared Fluorescent Cell-Based Assays. LI-COR Biosciences.
- Hoffmann, G., et al. (2008). ). A functional siRNA screen for novel regulators of mTORC1 signaling. Poster presented at ASCB Annual Meeting.
- Voets, O., Tielen, F., Elstak, E., Benschop, J., Grimbergen, M., Stallen, J., et al. (2017). Highly efficient gene inactivation by adenoviral CRISPR/Cas9 in human primary cells. PLoS ONE, 12(8). e0182974. DOI: 10.1371/journal.pone.0182974